Biological Chemistry
Staffs
Shin-ichi Hoshino, Ph.D., Professor
Tsuyoshi Udagawa, Ph.D., Associate Professor
Hiroto Inagaki, Ph.D., Research Associate
Tsuyoshi Udagawa, Ph.D., Associate Professor
Hiroto Inagaki, Ph.D., Research Associate
Research Project
- Mechanism of mRNA decay in eukaryotes
- Posttranscriptional regulation of gene expression
- Mechanism of mRNA quality control (surveillance)
- Roles of eRF3 family G proteins in the regulation of mRNA decay
Our laboratory conducts advanced research to gain a better understanding of the fundamental mechanisms of gene expression and RNA metabolism using genetic, molecular and biochemical approaches. We also focus upon the roles of RNA metabolism in cancer, apoptosis, aging, development and genetic diseases. The issues are being investigated in systems ranging from yeast to human cells.
Our efforts are currently devoted to understanding the molecular mechanism triggering mRNA decay. In eukaryotes, poly(A)-tail shortening is the rate-limiting step in the degradation of most mRNAs, and two major mRNA deadenylase complexes, Caf1-Ccr4 and Pan2-Pan3, play central roles in this process, referred to as deadenylation. Previoiusly, we have demonstrated that translation termination factor eRF3 mediates deadenylation and decay of mRNA in a manner coupled to translation termination. Based on our recent work in yeast and mammal, we have proposed an initiation mechanism of mRNA degradation as described in Figure (3) (see below).
We have also demonstrated that tumor suppressor Tob mediates activation of mRNA deadenylase.
(1) How mRNA is degraded after completion of gene expression.
Our efforts are currently devoted to understanding the molecular mechanism triggering mRNA decay. In eukaryotes, poly(A)-tail shortening is the rate-limiting step in the degradation of most mRNAs, and two major mRNA deadenylase complexes, Caf1-Ccr4 and Pan2-Pan3, play central roles in this process, referred to as deadenylation. Previoiusly, we have demonstrated that translation termination factor eRF3 mediates deadenylation and decay of mRNA in a manner coupled to translation termination. Based on our recent work in yeast and mammal, we have proposed an initiation mechanism of mRNA degradation as described in Figure (3) (see below).
We have also demonstrated that tumor suppressor Tob mediates activation of mRNA deadenylase.
(1) How mRNA is degraded after completion of gene expression.
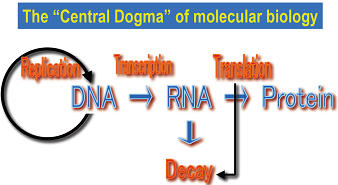
After (or during) translation, mRNA is degraded. However, it has not been elucidated what triggers mRNA decay.
(2) mRNA decay pathway; deadenylation is the first and late-limiting step in degradation of most mRNAs.
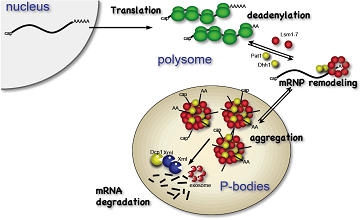
During and after translation, poly(A) tails of mRNA is gradually shortened.
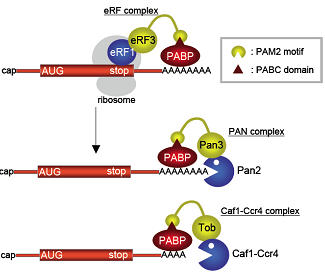
(3) Proposed model of mRNA deadenylation.
Translation termination complex eRF1-eRF3, Pan2-Pan3, and Caf1-Ccr4 competitively interact with polyadenyate-binding protein PABPC1. In each complex, eRF3, Pan3, and Tob, respectively, mediate PABPC1 binding, and a combination of a PAM2 motif and a PABC domain is commonly utilized for their contacts. A translation-dependent exchange of eRF1-eRF3 for the deadenylase occurs on PABPC1. Consequently, PABPC1 binding leads to the activation of Pan2-Pan3 and Caf1-Ccr4.
Translation termination complex eRF1-eRF3, Pan2-Pan3, and Caf1-Ccr4 competitively interact with polyadenyate-binding protein PABPC1. In each complex, eRF3, Pan3, and Tob, respectively, mediate PABPC1 binding, and a combination of a PAM2 motif and a PABC domain is commonly utilized for their contacts. A translation-dependent exchange of eRF1-eRF3 for the deadenylase occurs on PABPC1. Consequently, PABPC1 binding leads to the activation of Pan2-Pan3 and Caf1-Ccr4.
Contact Information
Department of Biological Chemistry,
Graduate School of Pharmaceutical Sciences, Nagoya City University,
3-1, Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan
E-mail:hoshino@phar.nagoya-cu.ac.jp
TEL/FAX:+81-52-836-3427
Graduate School of Pharmaceutical Sciences, Nagoya City University,
3-1, Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan
E-mail:hoshino@phar.nagoya-cu.ac.jp
TEL/FAX:+81-52-836-3427

